Research Goals
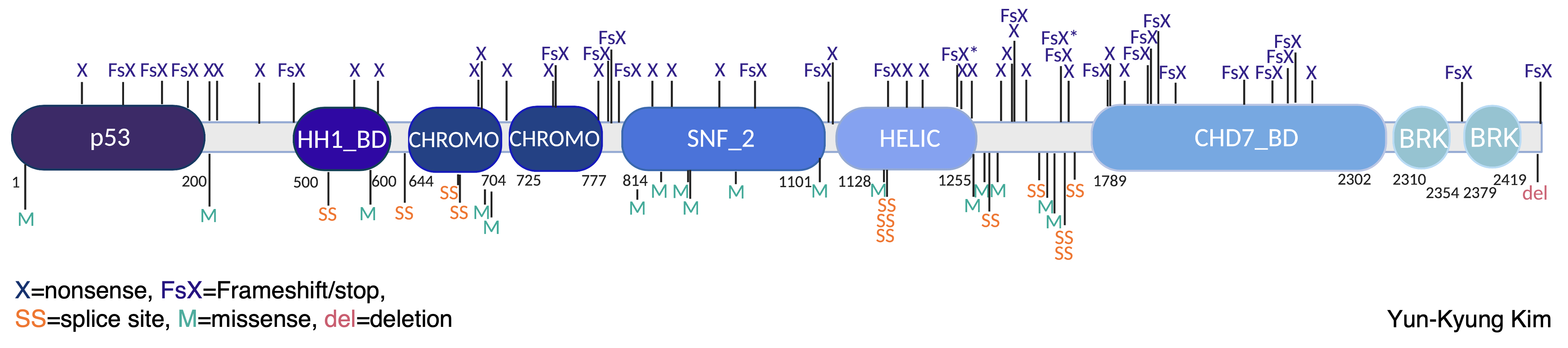
Disruptive mutations in the gene CHD8 (Chromodomain Helicase DNA-binding protein 8) are highly associated with a diagnosis of autism spectrum disorder (ASD) and other neurodevelopmental disorders. These changes often result in the disruption of one copy of CHD8, leading to less CHD8 protein expression in developing tissues such as the neocortex.
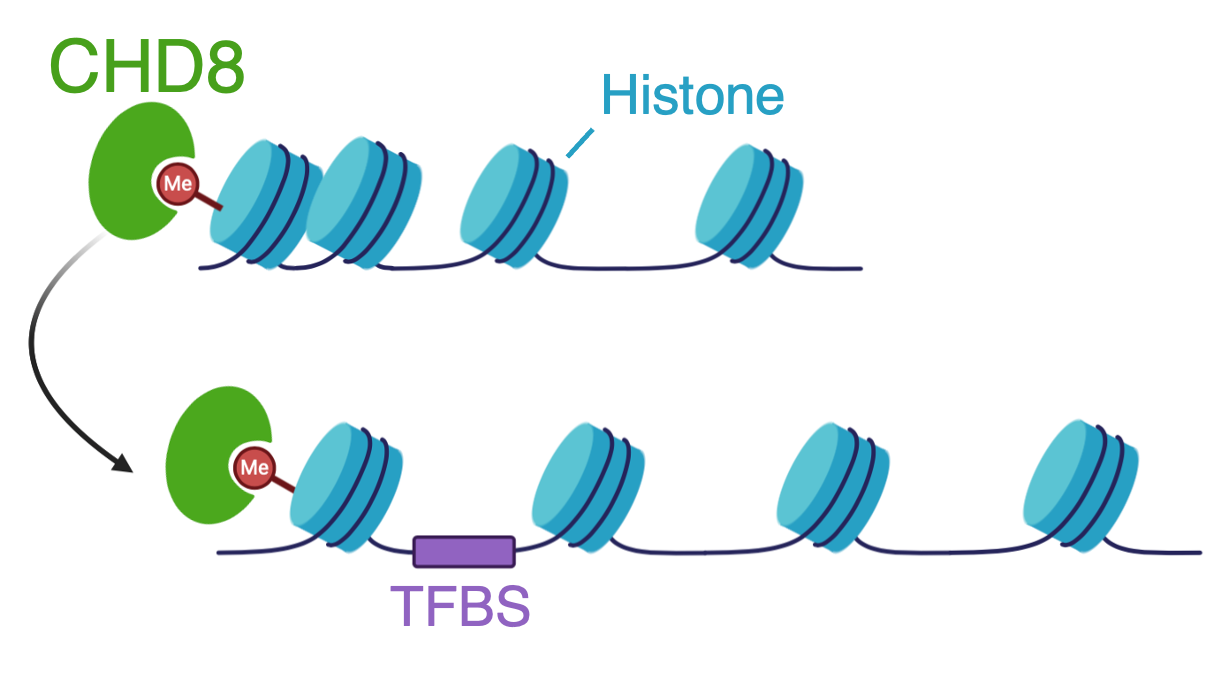
CHD8 binds to histone 3 lysine 4 (H3K4) methyl groups and remodels (or moves) nucleosomes along the DNA. By changing the accessibility of genomic regions, important regulatory regions may be revealed, such as transcription factor binding sites (TFBS) at promoter sites. In prior studies with the Noonan lab at Yale University, we performed chromatin-immunopreciptation assays followed by high-throughput sequencing (ChIP-seq), to determine the localization of CHD8 in neural stem cells, mouse cortex, and prenatal human brain. We found that CHD8 binds most often to promoter regions of active genes. CHD8 consistently bound gene targets across samples, including human neural stem cells (hNSCs), mouse cortex, and prenatal human brain.
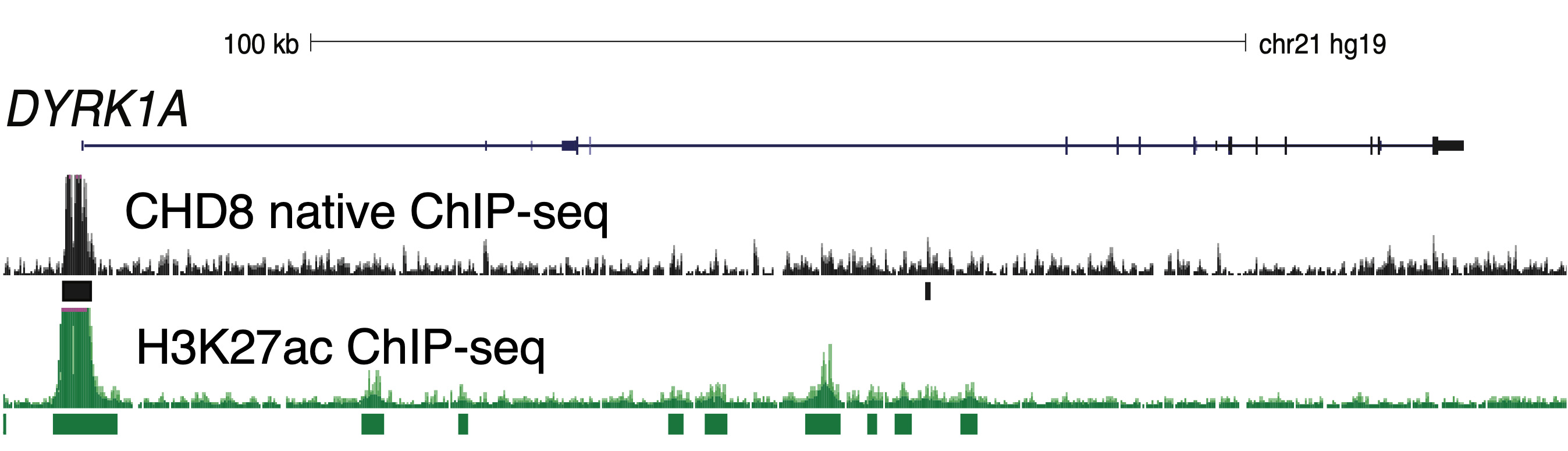
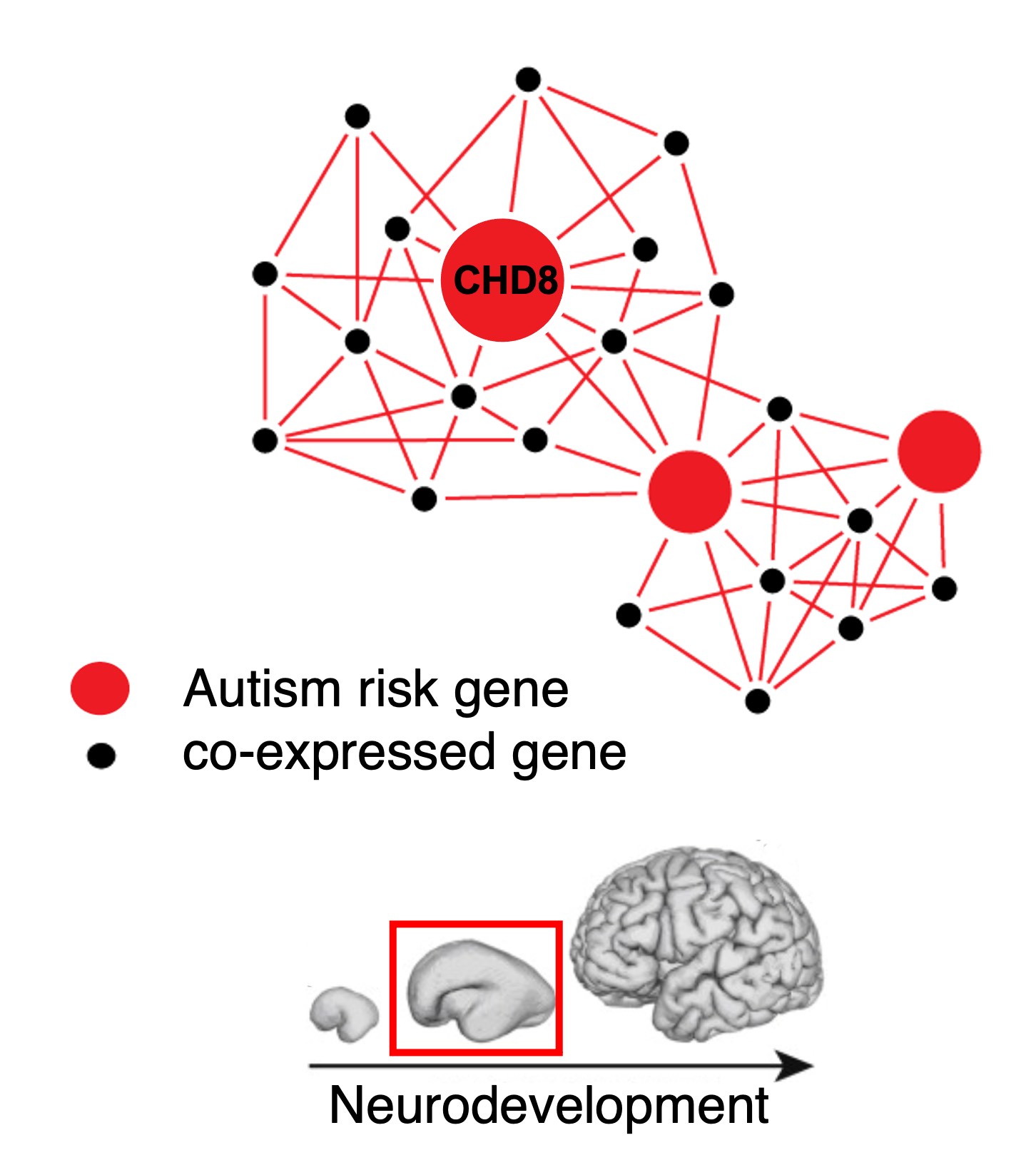
Additionally, we found that CHD8 regulates the expression of other genes associated with the likelihood of autism during neurodevelopment, and that these genes are co-expressed in networks that are also associated with likelihood of autism. Disruption of CHD8 function may therefore alter the expression of the genes in these networks. Our research aims to characterize the molecular and circuit-level changes associated with loss of CHD8 function. Specifically, we seek to understand how loss of CHD8 affects genes, cell types, and regulatory networks critical for neurodevelopment.
Research Projects
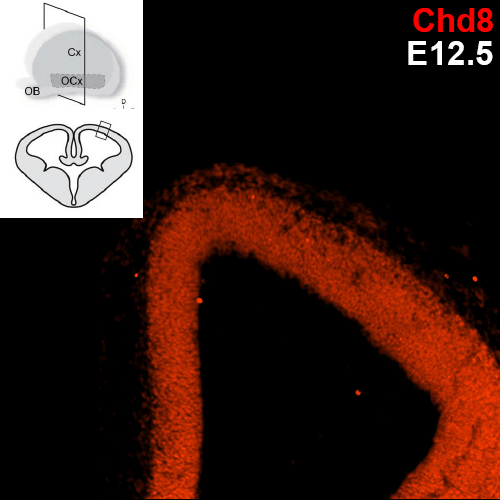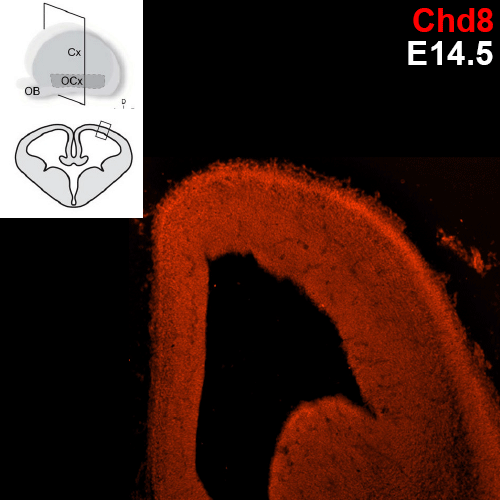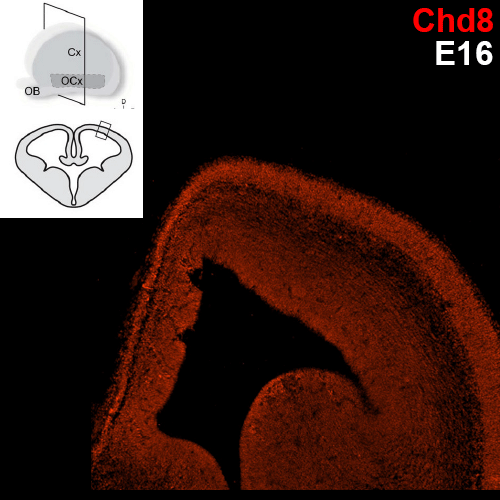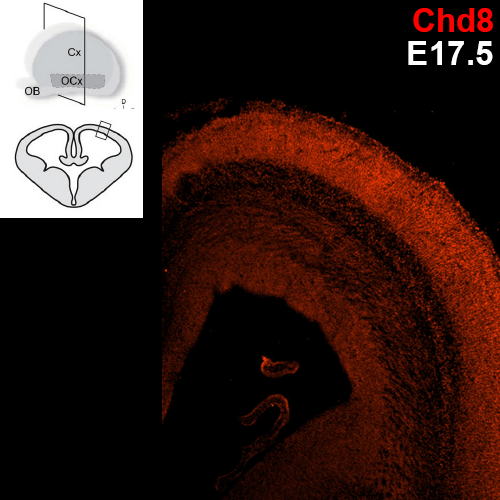
Which specific cell types show Chd8 expression?
Previous research conducted with the Noonan lab found regional expression of Chd8 is dynamic with embryonic age in mice. The uniform expression across the neocortex at E12.5 gives way to increasingly regionalized expression in the mature neurons of the developing mouse cortex, including cells of the cortical plate, the subplate, and the mantle zone. We are employing single cell analysis in developmental timepoints spanning corticogenesis, using a heterozygous Chd8+/- mouse line to determine the genome-wide expression and epigenetic profiles that define distinct brain cell populations in the context of Chd8 loss. Additionally, we are examining a role for Chd8 in non-cortical brain structures.
Which genes are regulated by Chd8 in specific cell types?
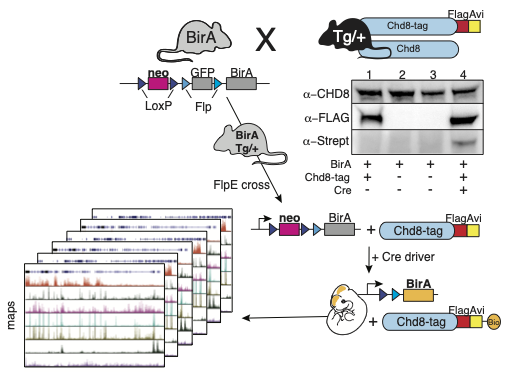
We have generated a transgenic Chd8-Avi-tagged mouse line that allows for cell-type specific biotinylation by a Cre-activated biotin ligase, BirA. We are performing streptavidin-mediated ChIP-seq (bioChIP-seq) in tissues expressing activated BirA in cell-types of interest using Cre driver lines to target biotinylation. This will generate chromatin maps of distinct cell types to assess genes and biological pathways regulated by Chd8.
CHD8 function in hNSCs, a model of early neurodevelopment
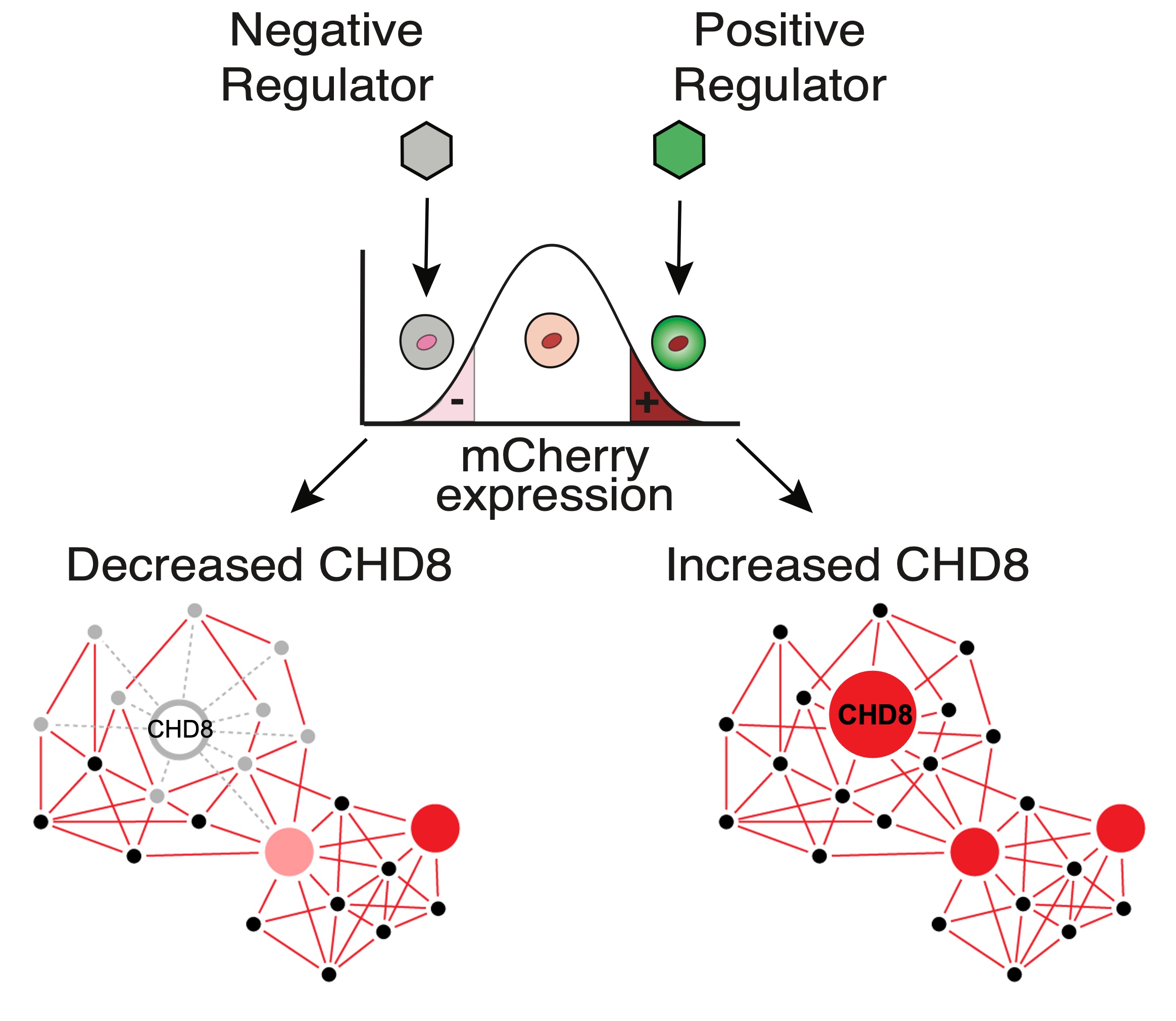
What are the negative and positive regulators of CHD8 expression? We are studying the effects of bioactive compounds such as hormones on CHD8 expression by using an mCherry-tagged CHD8 reporter in hNSCs.

A long-term consequence of Chd8 haploinsufficiency: Spontaneous seizures in Chd8+/- mice
Some people who carry a CHD8 mutation have a history of seizures or abnormal EEG, but this potential co-morbidity is not well characterized in human cohorts or model systems. We have found that seizure susceptibility increases with age in Chd8+/- mice. What are the brain regions and circuits responsible for these seizures? Identifying molecular markers preceding abnormal brain activity in our Chd8+/- mice may help to understand the etiology and progression of the seizure phenotype seen in patients with damaging CHD8 mutations.